Browse by Organisms Motifs
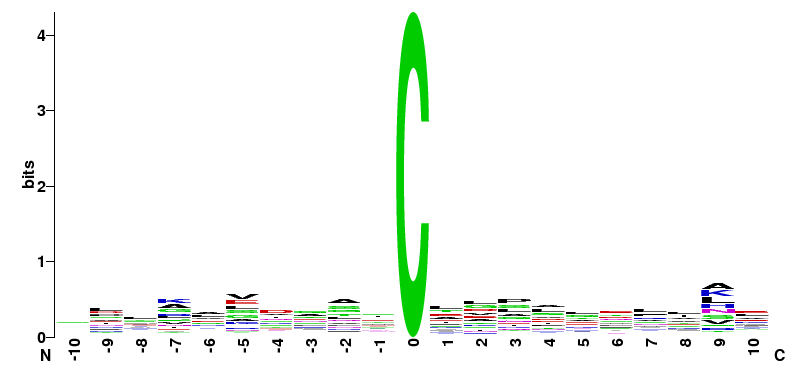
| R. norvegicus Motifs of RC04 group | ||||
 |
| No. | SwissProt ID | Position | S-nitrosylated Peptide | PubMed ID |
| 1 | 1433Z_RAT | 25 | QAERYDDMAA C MKSVTEQGAE | 17629318 |
| 2 | AATC_RAT | 83 | PILGLAEFRS C ASQLVLGDNS | 22178444 |
| 3 | AATC_RAT | 83 | PILGLAEFRS C ASQLVLGDNS | 19101475 |
| 4 | ABCC8_RAT | 717 | QLTMIVGQVG C GKSSLLLATL | 22178444 |
| 5 | ABCC8_RAT | 717 | QLTMIVGQVG C GKSSLLLATL | 19284878 |
| 6 | ARFG1_RAT | 350 | AKSPSSDSWT C ADASTGRRSS | 16418269 |
| 7 | ARRB2_RAT | 410 | MKDDDCDDQF C | 22178444 |
| 8 | ARRB2_RAT | 410 | MKDDDCDDQF C | 18691971 |
| 9 | AT1A3_RAT | 364 | VETLGSTSTI C SDKTGTLTQN | 16418269 |
| 10 | AT2B2_RAT | 497 | CETMGNATAI C SDKTGTLTTN | 22178444 |
| 11 | AT2B2_RAT | 497 | CETMGNATAI C SDKTGTLTTN | 16418269 |
| 12 | AT2B2_RAT | 571 | LPRQVGNKTE C GLLGFVLDLR | 22178444 |
| 13 | AT2B2_RAT | 571 | LPRQVGNKTE C GLLGFVLDLR | 16418269 |
| 14 | CAP1_RAT | 92 | ERALLVTASQ C QQPAGNKLSD | 22178444 |
| 15 | CAP1_RAT | 92 | ERALLVTASQ C QQPAGNKLSD | 19101475 |
| 16 | CDK17_RAT | 233 | IRLEHEEGAP C TAIREVSLLK | 22178444 |
| 17 | CDK17_RAT | 233 | IRLEHEEGAP C TAIREVSLLK | 16418269 |
| 18 | CH60_RAT | 442 | VEEGIVLGGG C ALLRCIPALD | 16418269 |
| 19 | CNGA2_RAT | 460 | TLKKVRIFQD C EAGLLVELVL | 22178444 |
| 20 | CNGA2_RAT | 460 | TLKKVRIFQD C EAGLLVELVL | 10809749 |
| 21 | CNGA2_RAT | 460 | TLKKVRIFQD C EAGLLVELVL | 15688001 |
| 22 | CPLX2_RAT | 90 | AEEKAALEQP C EGSLTRPKKA | 16418269 |
| 23 | CSK2B_RAT | 109 | EKYQQGDFGY C PRVYCENQPM | 21278135 |
| 24 | CSK2B_RAT | 114 | GDFGYCPRVY C ENQPMLPIGL | 21278135 |
| 25 | DCE2_RAT | 45 | KFTGGIGNKL C ALLYGDSEKP | 22178444 |
| 26 | DCE2_RAT | 45 | KFTGGIGNKL C ALLYGDSEKP | 16418269 |
| 27 | DPYL2_RAT | 504 | VPRGLYDGPV C EVSVTPKTVT | 22178444 |
| 28 | DPYL2_RAT | 504 | VPRGLYDGPV C EVSVTPKTVT | 16418269 |
| 29 | DPYL2_RAT | 504 | VPRGLYDGPV C EVSVTPKTVT | 17629318 |
| 30 | EAA2_RAT | 562 | TLAANGKSAD C SVEEEPWKRE | 22178444 |
| 31 | EAA2_RAT | 562 | TLAANGKSAD C SVEEEPWKRE | 16418269 |
| 32 | EIF3D_RAT | 258 | TDAILATLMS C TRSVYSWDIV | 21278135 |
| 33 | ELOC_RAT | 11 | MDGEEKTYGG C EGPDAMYVKL | 21278135 |
| 34 | ENOG_RAT | 270 | DFKSPADPSR C ITGDQLGALY | 22178444 |
| 35 | ENOG_RAT | 270 | DFKSPADPSR C ITGDQLGALY | 19101475 |
| 36 | ENOG_RAT | 357 | IGSVTEAIQA C KLAQENGWGV | 22178444 |
| 37 | ENOG_RAT | 357 | IGSVTEAIQA C KLAQENGWGV | 19101475 |
| 38 | G3P_RAT | 23 | GRLVTRAAFS C DKVDIVAIND | 17629318 |
| 39 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 7902582 |
| 40 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 8626764 |
| 41 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 15951807 |
| 42 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 16418269 |
| 43 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 18335467 |
| 44 | G3P_RAT | 150 | NSLKIVSNAS C TTNCLAPLAK | 19366988 |
| 45 | G3P_RAT | 154 | IVSNASCTTN C LAPLAKVIHD | 16418269 |
| 46 | G3P_RAT | 154 | IVSNASCTTN C LAPLAKVIHD | 18335467 |
| 47 | G3P_RAT | 154 | IVSNASCTTN C LAPLAKVIHD | 19366988 |
| 48 | GCYA3_RAT | 243 | SLMPPCFRSE C TEFVNQPYLL | 22178444 |
| 49 | GCYA3_RAT | 243 | SLMPPCFRSE C TEFVNQPYLL | 17636120 |
| 50 | GCYA3_RAT | 243 | SLMPPCFRSE C TEFVNQPYLL | 18669924 |
| 51 | GCYB1_RAT | 122 | YPGMRAPSFR C TDAEKGKGLI | 22178444 |
| 52 | GCYB1_RAT | 122 | YPGMRAPSFR C TDAEKGKGLI | 17636120 |
| 53 | GCYB1_RAT | 122 | YPGMRAPSFR C TDAEKGKGLI | 18669924 |
| 54 | GDIA_RAT | 317 | PIKNTNDANS C QIIIPQNQVN | 22178444 |
| 55 | GDIA_RAT | 317 | PIKNTNDANS C QIIIPQNQVN | 17629318 |
| 56 | GLNA_RAT | 269 | KAMREENGLR C IEEAIDKLSK | 22178444 |
| 57 | GLNA_RAT | 269 | KAMREENGLR C IEEAIDKLSK | 19101475 |
| 58 | GNAO_RAT | 140 | RLWGDSGIQE C FNRSREYQLN | 22178444 |
| 59 | GNAO_RAT | 140 | RLWGDSGIQE C FNRSREYQLN | 16418269 |
| 60 | GSTM1_RAT | 87 | MRYLARKHHL C GETEEERIRA | 22178444 |
| 61 | GSTM1_RAT | 87 | MRYLARKHHL C GETEEERIRA | 19101475 |
| 62 | GSTP1_RAT | 102 | MVNDGVEDLR C KYGTLIYTNY | 22178444 |
| 63 | GSTP1_RAT | 102 | MVNDGVEDLR C KYGTLIYTNY | 19101475 |
| 64 | HBB1_RAT | 94 | GTFAHLSELH C DKLHVDPENF | 19101475 |
| 65 | HBB2_RAT | 94 | GTFAHLSELH C DKLHVDPENF | 19101475 |
| 66 | HBB2_RAT | 126 | GHHLGKEFTP C AQAAFQKVVA | 19101475 |
| 67 | HMCS1_RAT | 417 | DLKSRLDSRT C VAPDVFAENM | 22178444 |
| 68 | HMCS1_RAT | 417 | DLKSRLDSRT C VAPDVFAENM | 19101475 |
| 69 | HPCA_RAT | 185 | SDPSIVRLLQ C DPSSASQF | 22178444 |
| 70 | HPCA_RAT | 185 | SDPSIVRLLQ C DPSSASQF | 19101475 |
| 71 | HSP74_RAT | 417 | NSPAEEGSSD C EVFPKNHAAP | 19101475 |
| 72 | K6PP_RAT | 81 | GSNIVEAKWE C VSSILQVGGT | 16418269 |
| 73 | KCRB_RAT | 254 | GNMKEVFTRF C TGLTQIETLF | 17629318 |
| 74 | KCRB_RAT | 254 | GNMKEVFTRF C TGLTQIETLF | 18335467 |
| 75 | KCRB_RAT | 254 | GNMKEVFTRF C TGLTQIETLF | 19366988 |
| 76 | KCRB_RAT | 283 | WNPHLGYILT C PSNLGTGLRA | 16418269 |
| 77 | KCRS_RAT | 317 | WNERLGYILT C PSNLGTGLRA | 22178444 |
| 78 | KCRS_RAT | 317 | WNERLGYILT C PSNLGTGLRA | 16418269 |
| 79 | KPYM_RAT | 49 | PITARNTGII C TIGPASRSVE | 22178444 |
| 80 | KPYM_RAT | 49 | PITARNTGII C TIGPASRSVE | 17629318 |
| 81 | KPYM_RAT | 49 | PITARNTGII C TIGPASRSVE | 18335467 |
| 82 | KPYM_RAT | 49 | PITARNTGII C TIGPASRSVE | 19366988 |
| 83 | KPYM_RAT | 326 | RCNRAGKPVI C ATQMLESMIK | 22178444 |
| 84 | KPYM_RAT | 326 | RCNRAGKPVI C ATQMLESMIK | 19101475 |
| 85 | KPYM_RAT | 358 | VANAVLDGAD C IMLSGETAKG | 22178444 |
| 86 | KPYM_RAT | 358 | VANAVLDGAD C IMLSGETAKG | 16418269 |
| 87 | KPYM_RAT | 423 | AMGSVEASYK C LAAALIVLTE | 22178444 |
| 88 | KPYM_RAT | 423 | AMGSVEASYK C LAAALIVLTE | 17629318 |
| 89 | KPYM_RAT | 474 | HLYRGIFPVL C KDAVLDAWAE | 22178444 |
| 90 | KPYM_RAT | 474 | HLYRGIFPVL C KDAVLDAWAE | 17629318 |
| 91 | MDHC_RAT | 137 | IVVGNPANTN C LTASKSAPSI | 22178444 |
| 92 | MDHC_RAT | 137 | IVVGNPANTN C LTASKSAPSI | 17629318 |
| 93 | MDHC_RAT | 137 | IVVGNPANTN C LTASKSAPSI | 18335467 |
| 94 | MDHC_RAT | 137 | IVVGNPANTN C LTASKSAPSI | 19366988 |
| 95 | MK03_RAT | 179 | KPSNLLINTT C DLKICDFGLA | 16418269 |
| 96 | MK08_RAT | 116 | IVMELMDANL C QVIQMELDHE | 11121042 |
| 97 | MYPR_RAT | 201 | SKTSASIGSL C ADARMYGVLP | 22178444 |
| 98 | MYPR_RAT | 201 | SKTSASIGSL C ADARMYGVLP | 16418269 |
| 99 | NAC2_RAT | 537 | FQDRLLHVSE C MGTVDVRVVR | 22178444 |
| 100 | NAC2_RAT | 537 | FQDRLLHVSE C MGTVDVRVVR | 16418269 |
| 101 | OGT1_RAT | 610 | NHFIDLSQIP C NGKAADRIHQ | 22178444 |
| 102 | OGT1_RAT | 610 | NHFIDLSQIP C NGKAADRIHQ | 16418269 |
| 103 | OTUB1_RAT | 91 | YIRKTRPDGN C FYRAFGFSHL | 21278135 |
| 104 | PARK7_RAT | 53 | PVQCSRDVVI C PDTSLEEAKT | 22178444 |
| 105 | PARK7_RAT | 53 | PVQCSRDVVI C PDTSLEEAKT | 16316629 |
| 106 | PARK7_RAT | 106 | ENRKGLIAAI C AGPTALLAHE | 22178444 |
| 107 | PARK7_RAT | 106 | ENRKGLIAAI C AGPTALLAHE | 16418269 |
| 108 | PARK7_RAT | 121 | ALLAHEVGFG C KVTSHPLAKD | 22178444 |
| 109 | PARK7_RAT | 121 | ALLAHEVGFG C KVTSHPLAKD | 16418269 |
| 110 | PFKAP_RAT | 81 | GSNIVEAKWE C VSSILQVGGT | 22178444 |
| 111 | PGAM1_RAT | 153 | ADLTEDQLPS C ESLKDTIARA | 22178444 |
| 112 | PGAM1_RAT | 153 | ADLTEDQLPS C ESLKDTIARA | 19101475 |
| 113 | PGAM1_RAT | 153 | ADLTEDQLPS C ESLKDTIARA | 19483679 |
| 114 | PITM1_RAT | 259 | ARMLAQRMAK C NTGSEGPEAQ | 22178444 |
| 115 | PITM1_RAT | 259 | ARMLAQRMAK C NTGSEGPEAQ | 19101475 |
| 116 | PLEC1_RAT | 4316 | QTYLELSEQE C EWEEITISSS | 17224453 |
| 117 | PP1R7_RAT | 112 | SLCLRQNLIK C IENLDELQSL | 22178444 |
| 118 | PP1R7_RAT | 112 | SLCLRQNLIK C IENLDELQSL | 19101475 |
| 119 | PPIA_RAT | 161 | KTSKKITISD C GQL | 22178444 |
| 120 | PPIA_RAT | 161 | KTSKKITISD C GQL | 17629318 |
| 121 | Q9QX80_RAT | 104 | YFTKFGEVVD C TIKMDPNTGR | 22178444 |
| 122 | Q9QX80_RAT | 104 | YFTKFGEVVD C TIKMDPNTGR | 14722087 |
| 123 | RIPL1_RAT | 47 | EFERVIDQHG C EAIARLMPKV | 22178444 |
| 124 | RIPL1_RAT | 47 | EFERVIDQHG C EAIARLMPKV | 19607794 |
| 125 | RS4X_RAT | 41 | PSTGPHKLRE C LPLIIFLRNR | 19101475 |
| 126 | RTN3_RAT | 47 | ALGAKSCGSS C AVGLSSLCSD | 22178444 |
| 127 | RTN3_RAT | 47 | ALGAKSCGSS C AVGLSSLCSD | 16418269 |
| 128 | RTN4_RAT | 403 | ANVESKVDRK C LEDSLEQKSL | 19101475 |
| 129 | SERA_RAT | 369 | QGTSLKNAGT C LSPAVIVGLL | 16418269 |
| 130 | STK39_RAT | 533 | KTLTFKLASG C DGAEIPDEVK | 22178444 |
| 131 | STK39_RAT | 533 | KTLTFKLASG C DGAEIPDEVK | 19101475 |
| 132 | SYT2_RAT | 293 | YVPTAGKLTV C ILEAKNLKKM | 22178444 |
| 133 | SYT2_RAT | 293 | YVPTAGKLTV C ILEAKNLKKM | 16418269 |
| 134 | TBA1A_RAT | 295 | QLSVAEITNA C FEPANQMVKC | 22178444 |
| 135 | TBA1A_RAT | 295 | QLSVAEITNA C FEPANQMVKC | 16418269 |
| 136 | TBA1A_RAT | 315 | CDPRHGKYMA C CLLYRGDVVP | 22178444 |
| 137 | TBA1A_RAT | 315 | CDPRHGKYMA C CLLYRGDVVP | 17629318 |
| 138 | TBA1A_RAT | 316 | DPRHGKYMAC C LLYRGDVVPK | 22178444 |
| 139 | TBA1A_RAT | 316 | DPRHGKYMAC C LLYRGDVVPK | 17629318 |
| 140 | TBA1A_RAT | 376 | GDLAKVQRAV C MLSNTTAIAE | 22178444 |
| 141 | TBA1A_RAT | 376 | GDLAKVQRAV C MLSNTTAIAE | 17629318 |
| 142 | TBA1B_RAT | 295 | QLSVAEITNA C FEPANQMVKC | 16418269 |
| 143 | TBA1B_RAT | 376 | GDLAKVQRAV C MLSNTTAIAE | 16418269 |
| 144 | TBB5_RAT | 12 | REIVHIQAGQ C GNQIGAKFWE | 16418269 |
| 145 | TBB5_RAT | 239 | VSATMSGVTT C LRFPGQLNAD | 16418269 |
| 146 | TBB5_RAT | 354 | WIPNNVKTAV C DIPPRGLKMA | 16418269 |
| 147 | TEBP_RAT | 40 | NFEKSKLTFS C LGGSDNFKHL | 21278135 |
| 148 | TEBP_RAT | 58 | KHLNEIDLFH C IDPNDSKHKR | 21278135 |
| 149 | TPIS_RAT | 21 | GNWKMNGRKK C LGELICTLNA | 16418269 |
| 150 | TPIS_RAT | 27 | GRKKCLGELI C TLNAAKLPAD | 16418269 |
| 151 | TPIS_RAT | 67 | DPKIAVAAQN C YKVTNGAFTG | 19101475 |
| 152 | TRPV1_RAT | 621 | STPHKCRGSA C KPGNSYNSLY | 22178444 |
| 153 | TRPV1_RAT | 621 | STPHKCRGSA C KPGNSYNSLY | 16998480 |
| 154 | UB2D2_RAT | 85 | HPNINSNGSI C LDILRSQWSP | 21278135 |
| 155 | UCHL1_RAT | 152 | AHDSVAQEGQ C RVDDKVNFHF | 22178444 |
| 156 | UCHL1_RAT | 152 | AHDSVAQEGQ C RVDDKVNFHF | 16418269 |
| 157 | UCHL1_RAT | 152 | AHDSVAQEGQ C RVDDKVNFHF | 21278135 |
| 158 | UCHL1_RAT | 220 | GEVRFSAVAL C KAA | 21278135 |
| 159 | VATB2_RAT | 162 | DIMGQPINPQ C RIYPEEMIQT | 22178444 |
| 160 | VATB2_RAT | 162 | DIMGQPINPQ C RIYPEEMIQT | 16418269 |
| 161 | VISL1_RAT | 187 | SDPSIVLLLQ C DIQK | 16418269 |
